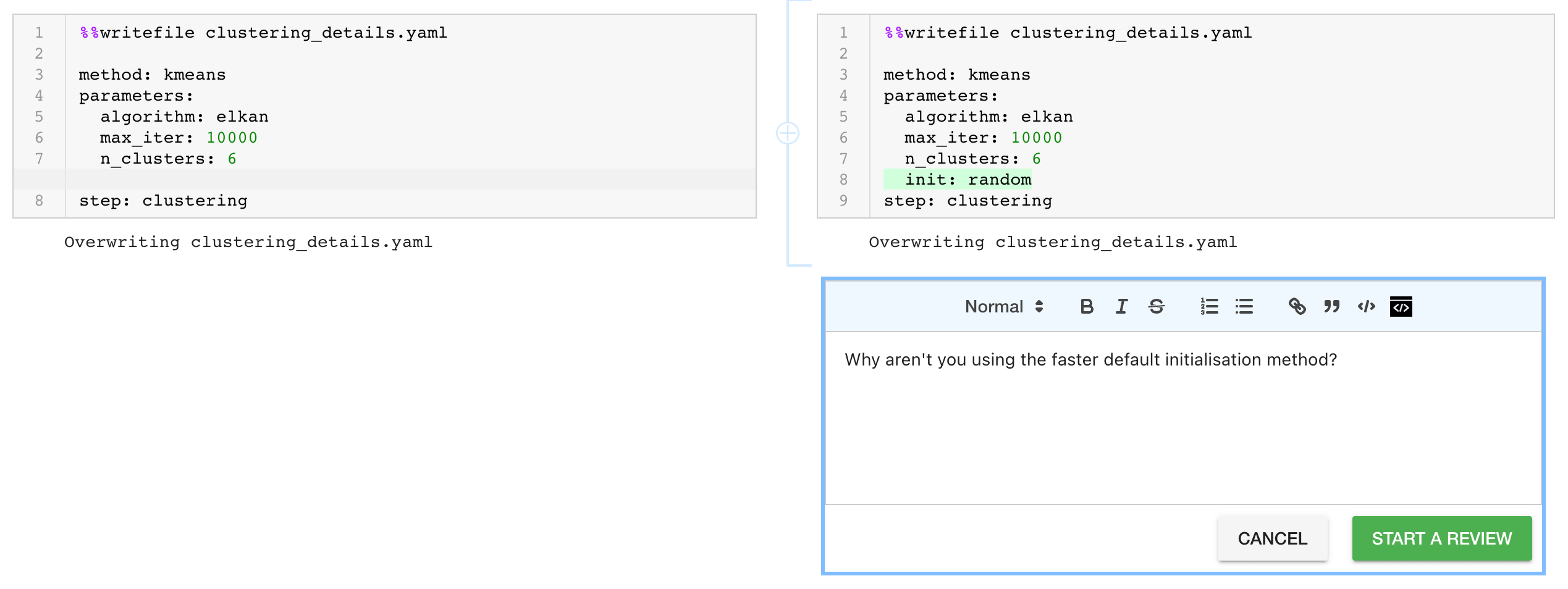
How To Run Yaml With Jupyter Notebook
You might use YAML in a Jupyter notebook to manage deployment configuration files or to automate and track machine learning experimentation, but YAML doesn’t have to be used for configuration only – in some cases, full datasets are stored in YAML format.
Let’s take a look at how you can work with YAML in Jupyter.
YAML parsing / writing tools
PyYAML
You can use the pyyaml package to read and write YAML using Python.
Let’s say you have a YAML config file transaction_clustering.yaml, and in it you’ve saved some of the steps you used in a data science experiment:
---
- step: scaling
method: robust
parameters:
with_centering: true
- step: clustering
method: kmeans
parameters:
n_clusters: 6
max_iter: 10000
algorithm: elkan
You can read the file in with the pyyaml shorthand functions load or safe_load. The safe_load function prevents the running arbitrary of Python code as the YAML is loaded, so it is usually preferred.
import yaml
experiment_details = yaml.safe_load(open('transaction_clustering.yaml'))
print(experiment_details)
[{'step': 'scaling',
'method': 'robust',
'parameters': {'with_centering': True}},
{'step': 'clustering',
'method': 'kmeans',
'parameters': {'n_clusters': 6, 'max_iter': 10000, 'algorithm': 'elkan'}}]
The data in the YAML file has been read into a Python list of dictionaries, which you can now use as normal:
print(experiment_details[1]['parameters']['max_iter'])
10000
You can also save Python objects to YAML strings:
print(yaml.safe_dump(experiment_details[1]))
method: kmeans
parameters:
algorithm: elkan
max_iter: 10000
n_clusters: 6
step: clustering
And as YAML files:
yaml.safe_dump(experiment_details[1], open('clustering_details.yaml', 'w'))
Notice here that we’re using the safe version of the dump operation, which just supports basic YAML tags.
The yaml shorthand functions do have some limited configurability. For example, you can configure the dumping functions to include document markers, which aren’t included by default:
print(yaml.safe_dump(experiment_details[1], explicit_start=True))
---
method: kmeans
parameters:
algorithm: elkan
max_iter: 10000
n_clusters: 6
step: clustering
If you’d like more configurability than the shorthand functions provide, you can use the pyyaml loader and dumper classes directly.
yamlmagic
Another convenient Python package for working with YAML is yamlmagic, which provides the magic to turn YAML text into Python objects.
For example, we can use the yamlmagic cell magic to turn this cell into a YAML interpreting cell:
%load_ext yamlmagic
%%yaml clustering_details
step: clustering
method: kmeans
parameters:
n_clusters: 6
max_iter: 10000
algorithm: elkan
We now have this data, inputted in YAML format, in the Python variable clustering_details:
print(clustering_details)
{'step': 'clustering',
'method': 'kmeans',
'parameters': {'n_clusters': 6, 'max_iter': 10000, 'algorithm': 'elkan'}}
Writefile
You can use the built-in writefile magic to save YAML-formatted data into a file.
For example:
%%writefile clustering_details.yaml
step: clustering
method: kmeans
parameters:
n_clusters: 6
max_iter: 10000
algorithm: elkan
Here we save our clustering parameter data into the YAML file clustering_details.yaml, which we can now use elsewhere. You could just use an editor to save these parameters into a file, but if you’re already doing analysis or modelling in a Jupyter notebook, it’s convenient to continue working in the notebook.
Jupyter and YAML use cases
Let’s take a look at some of the contexts in which data scientists and data engineers may come across YAML files.
Configuration management for DevOps, MLOps, and DataOps
YAML is used widely in DevOps, MLOps and DataOps for orchestration, deployment configuration, data pipeline configuration, and ML pipeline configuration.
The Seldon documentation uses the writefile Jupyter magic to save graph-level metadata. You may find yourself designing your Seldon graphs in a Jupyter notebook and setting up the YAML files there as part of your development and testing process.
You can also imagine opening a Kubernetes cronjob YAML file to check when the cronjob is run:
yaml.safe_load(open('daily_clustering.yaml'))
{'apiVersion': 'batch/v1',
'kind': 'CronJob',
'metadata': {'name': 'clustering-daily:latest'},
'spec': {'schedule': '30 8 * * *',
'jobTemplate': {'spec': {'template': {'spec': {'containers': [{'name': 'clustering',
'image': 'clustering'}],
'command': ['run-clustering'],
'restartPolicy': 'OnFailure'}}}}}}
You could potentially also programmatically duplicate and edit these configuration files:
clustering_cron = yaml.safe_load(open("daily-clustering.yaml"))
for hour in (13, 14, 15, 16):
clustering_cron["spec"]["schedule"] = f"30 {hour} * * "
yaml.safe_dump(
clustering_cron, open(f"daily_clustering_{hour}h30m.yaml", "w"), sort_keys=False
)
Here we create four new cronjob definition files, which will run at 13:30, 14:30, 15:30, and 16:30 respectively.
Experiment automation and tracking
Data science preprocessing pipelines and machine learning models usually have many parameters and hyperparameters. YAML can be used to track the parameters used in individual experiments, or as part of hyperparameter tuning steps where modelling is performed and evaluated for many configurations.
Storing these kinds of parameters and hyperparameters in YAML has several benefits: It makes them human readable and easily human editable, it allows them to be version controlled, and it decouples parameters from code, so that code can be re-run on different parameter sets, enabling automation.
For experimentation tracking, you could use YAML files to track parameters of experiments in a version controlled folder:
transaction_clustering
|_ eda.ipynb
|_ transaction_clustering.ipynb
|_ experiment_parameters_20220404.yaml
|_ experiment_parameters_20220408.yaml
Here each YAML file could contain parameters for a particular version of your experiment:
date: 2022-04-04
data: iris.csv
clustering:
method: kmeans
fields:
- sepal_length
- sepal_width
- petal_length
- petal_width
parameters:
n_clusters: 3
max_iter: 100
algorithm: elkan
random_state: 22
Then this configuration YAML would be read in and used as variables in your modelling code in transaction_clustering.ipynb:
import pandas as pd
from sklearn.cluster import KMeans
import yaml
config = yaml.safe_load(open('experiment_parameters_20220404.yaml'))
print(config['date'])
df = pd.read_csv(config['data'])
clustering_config = config['clustering']
if clustering_config['method'] == 'kmeans':
kmeans_parameters = clustering_config['parameters']
clustering = KMeans(
n_clusters=kmeans_parameters['n_clusters'],
max_iter=kmeans_parameters['max_iter'],
algorithm=kmeans_parameters['algorithm'],
random_state=kmeans_parameters['random_state']
)
y = clustering.fit_predict(df[clustering_config['fields']])
Note how easy it would be to re-run the experiment set up in transaction_clustering.ipynb
using a different YAML config file with different parameters. You could run this experiment using many different
config files, and potentially different datasets, and at the end of each notebook save the results of the experiment to a file.
A similar approach can be used for config files for hyperparameter searches. For example, the hyperparameter tuning tool Google AI Platform can use YAML files to define the hyperparameter search space for use with various machine learning packages.
Data Storage
YAML can also be used to store data. Like JSON, it’s a useful format for nested data that can’t be represented well in a tabular format like CSV.
As an example, let’s have a look at some cricket match data provided by Cricsheet. First we download Indian Premier League data from the matches page and read it in:
ipl_data = yaml.safe_load(open('ipl/1082591.yaml'))
print(ipl_data.keys())
dict_keys(['meta', 'info', 'innings'])
We can navigate the data to the results of each ball in each innings:
ipl_data['innings'][0]
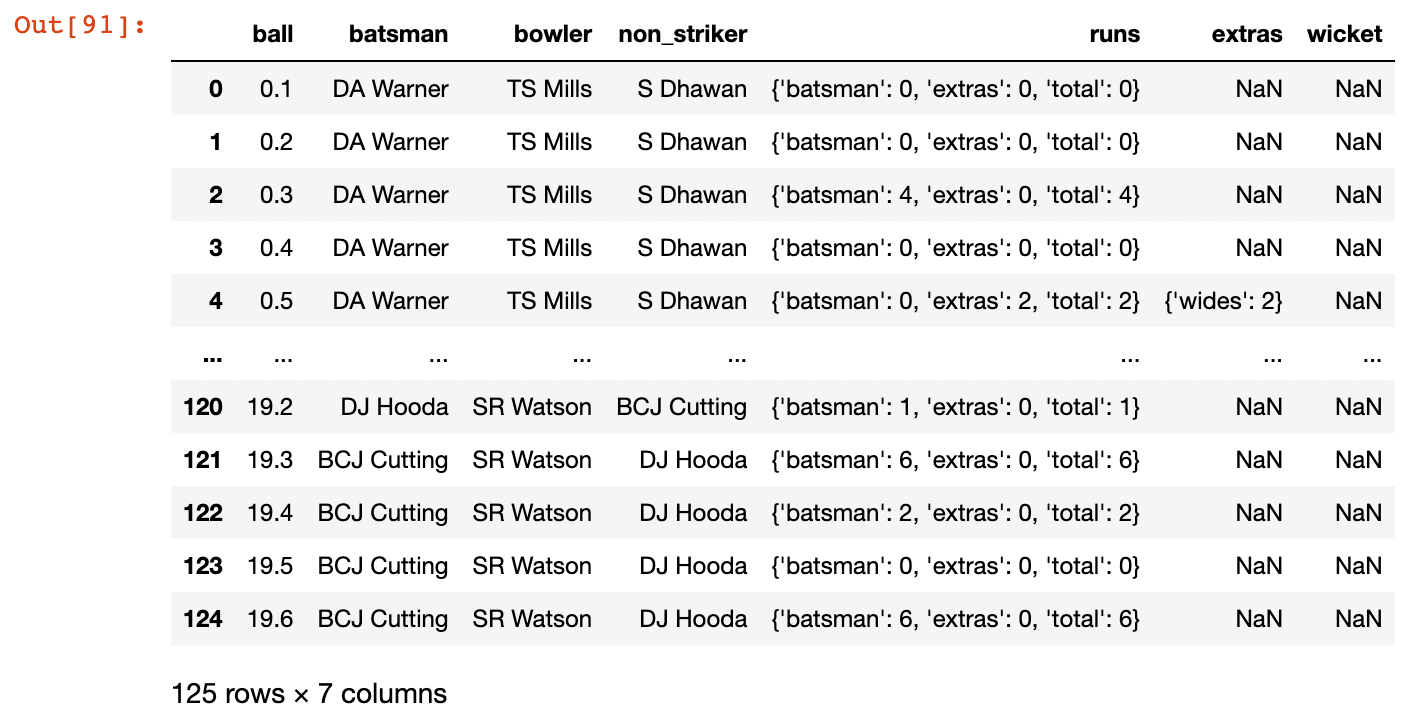
Then we can do some munging of the data into a format readable by pandas:
import pandas as pd
flattened_innings = [{**{'ball': k}, **v} for c in ipl_data['innings'][0]['1st innings']['deliveries'] for k, v in c.items()]
df = pd.DataFrame(flattened_innings)
print(df.head())
Using YAML outside of a Jupyter Notebook
We’ve looked at a couple of Python modules that allow us to create, edit, and import data from YAML files within a notebook. But often it’s more convenient to work with YAML outside of a Jupyter notebook.
Text Editors
If you’re already working in a Jupyter or Jupyter Lab environment, it’s easy enough to open a text file within the environment and create or edit your YAML there.
But you don’t have to stay in Jupyter – widely used editors like Visual Studio Code and Sublime Code let you create and edit YAML files, and also come with syntax highlighting that make YAML even easier to read. Many editors also have more advanced YAML plugins available, that work with specific flavours of YAML, and offer auto-completion and syntax verification. If you often work with large YAML files, this can be invaluable.
Terminal-based editors like Vim, Emacs, and Notepad all work as YAML editors too. If you need to log in to remote machines to check or edit YAML, you will likely need to use one of these.
yamllint
Linters are programs that check the correctness of your code. yamllint is a linter for YAML that checks if your YAML is syntactically correct and also does some cosmetic checking (for trailing whitespace, for example). It is worth running it on your YAML files before using them or committing them to a version control system.
Say we have an indentation error in our YAML:
---
- step: scaling
method: robust
parameters:
with_centering: true
- step: clustering
method: kmeans
parameters:
n_clusters: 6
max_iter: 10000
algorithm: elkan
That isn’t an easy error to spot by eye. The larger or more complex a YAML file is, the more difficult it is to pick up errors, so the automated checking done by linters can save time.
yamllint is a Python package that can be installed with pip, and then run from the command line. If we run it for the YAML above, it tells us about the error:
$ yamllint transaction_clustering_with_error.yaml
transaction_clustering_with_error.yaml
5:25 error trailing spaces (trailing-spaces)
8:10 error syntax error: mapping values are not allowed here (syntax)
It also picked up a trailing whitespace character in line 5, something we wouldn’t have noticed otherwise.
Sometimes yamllint is more strict than you may want it to be. For example, it will create an error if you leave out the document start marker --- in the first line of a YAML file. This can clash with the default dump behavior of pyyaml, which omits the start marker. But the yamllint configuration is adjustable through a custom configuration file, so you can set it to ignore rules you don’t want to be checked.
If you want yamllint to pass YAML created by the default pyyaml dump and safe_dump functions, you can use the following custom configuration:
rules:
document-start: disable
indentation:
indent-sequences: false
Without this config, running yamllint on one of our clustering_config cronjob files will fail:
$ yamllint daily_clustering_16h30mb.yaml
daily_clustering_16h30mb.yaml
1:1 warning missing document start "---" (document-start)
12:11 error wrong indentation: expected 12 but found 10 (indentation)
15:11 error wrong indentation: expected 12 but found 10 (indentation)
If we use the config file, these warnings and errors won’t be raised:
$ yamllint daily_clustering_16h30mb.yaml -c yamllint_config.yaml
How permissive or conservative you want your linting to be is up to you, and can depend on the applications you are using your YAML with.
Command line YAML navigators
The command line tools yq and shyaml allow you to navigate YAML from the command line. Let’s first look at yq:
$ yq . transaction_clustering.yaml
This prints the full contents of our YAML file. To pull out elements of a list, we use indices:
$ yq '.[0]' transaction_clustering.yaml
{
"step": "scaling",
"method": "robust",
"parameters": {
"with_centering": true
}
}
And we use dots to navigate down levels:
$ yq '.[1].method' transaction_clustering.yaml
"kmeans"
While yq is a Python package installable with pip, it is actually a wrapper for the JSON processor jq which you will have to install too. Installation instructions can be found on the yq PyPI site.
An alternative to yq is shyaml, which is also a pip-installable Python package. The shyaml script only reads from stdin (unlike yq, which can read from stdin and from files), so the syntax to use is:
$ cat transaction_clustering.yaml | shyaml get-value '0.method'
robust
$ cat transaction_clustering.yaml | shyaml get-value '1.parameters.n_clusters'
6
$ cat transaction_clustering.yaml | shyaml get-type '1.parameters.n_clusters'
int
Round up
Jupyter Notebooks is a convenient and interactive way for data scientists to interact with YAML, but one of Jupyter’s downsides is that it’s tricky to review.
ReviewNB solves this problem by supporting notebook diffs and comment conversation threads, which helps us spot problems (easy to make mistakes with nested YAML!) and enhances collaboration on notebook development.